
I was told when I first started blogging here at 10,000 Birds that I was never to use the short form, “10K.” But here I’m using it because someone ELSE used it … the Bird 10K project is an effort to do the whole DNA thing they do on groups of species on the whole mess of 10K (or more) birds. This is interesting right now because the AVian Phylogenomics Consortium has just announced the Bird 10K project, which ties together a pile of previously done research with some exciting new projects just taking off now.
From the Bird 10K project:
The Bird 10,000 genomes (B10K) project is an initiative to generate representative draft genome sequences from all extant bird species within the next five years. The establishment of this project is built on the success of the previous ordinal level project (http://avian.genomics.cn/en/index.html), which provided the first proof of concept for carrying out large-scale sequencing of multiple representative species across a vertebrate class and a window into the types of discoveries that can be made with such genomes…
The extraordinary diversity of Galapagos finches discovered during the 1826 voyage of the H.M.S. Beagle had an enormous impact on Darwin’s thinking about ‘On the Origin of Species’. His later studies on domesticated pigeons further inspired the development of the theory of evolution. Since then, bird studies have led to numerous pioneering findings that have established many new disciplines in biology, including Wallace’s biogeography, Mayr’s synthesis of speciation, MacArthur and Wilson’s island biogeography, Tinbergen’s ethology, and Hamilton’s kinship theory. With about 10,500 living species, birds constitute the most speciose class of terrestrial vertebrates and are adapted to and distributed throughout most terrestrial and shallow aquatic biomes on our planet. Given these attributes, birds continue to be widely used as models for studies on population genetics, neurobiology, development and animal conservation. …
…The phylogeny of birds has been one of the most challenging vertebrate groups to decipher. Recently, our ordinal level project, a large international consortium led by researchers from BGI, University of Copenhagen, and Duke University, and including investigators from more than 20 countries, sequenced and/or collected the genomes from 48 bird species representing nearly all orders and covering a broad range of evolutionary diversity. The consortium was able to resolve much of the contested phylogenetic history of modern avian orders…
Just to give you an idea of how cool this is, here is the “genome-scale phylogeny of birds” from “Whole-genome analyses resolve early branches in the tree of life of modern birds” in Science.
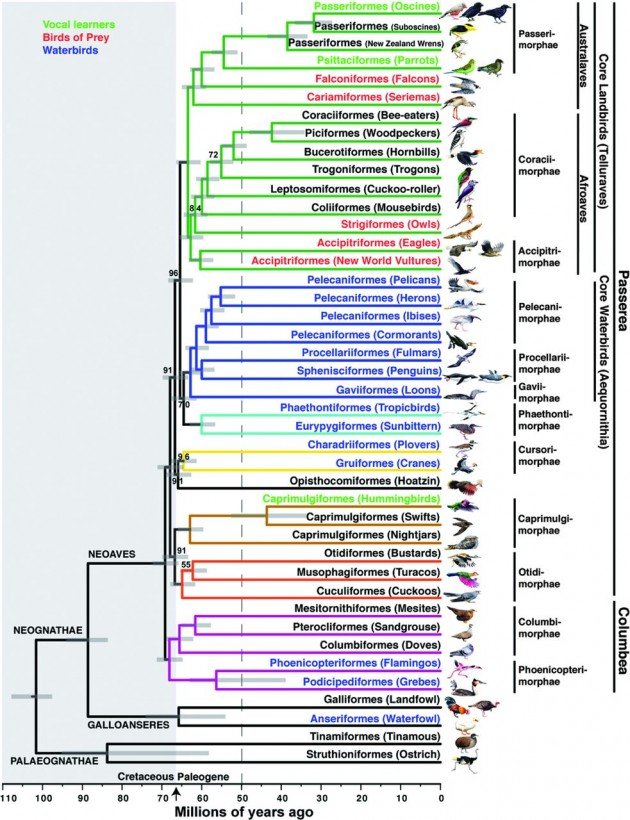
Fig. 1 Genome-scale phylogeny of birds.
The dated TENT inferred with ExaML. Branch colors denote well-supported clades in this and other analyses. All BS values are 100% except where noted. Names on branches denote orders (-iformes) and English group terms (in parentheses); drawings are of the specific species sequenced (names in table S1 and fig. S1). Order names are according to (36, 37) (SM6). To the right are superorder (-imorphae) and higher unranked names. In some groups, more than one species was sequenced, and these branches have been collapsed (noncollapsed version in fig. S1). Text color denotes groups of species with broadly shared traits, whether by homology or convergence. The arrow indicates the K-Pg boundary at 66 Ma, with the Cretaceous period shaded at left. The gray dashed line represents the approximate end time (50 Ma) by which nearly all neoavian orders diverged. Horizontal gray bars on each node indicate the 95% credible interval of divergence time in millions of years.











Leave a Comment